大胆设想,严谨求证
坚持创新,务实求真
重视人才发展,重铸科学之魂
Liang, Zhuobin
梁卓斌
Institute of Molecular Physiology
Junior Principal Investigator
zbliang@szbl.ac.cn
Timeline
-
2021 - Present
Shenzhen Bay Laboratory Junior Principal Investigator
-
2017 - 2021
Yale University Postdoctoral Associate
-
2015 - 2017
Yale School of Medicine Damon Runyon Postdoctoral Fellow
-
2009 - 2015
University of Michigan-Ann Arbor Ph.D. in Molecular Biology
-
2004 - 2008
Shanghai Jiao Tong University B.S. in Plant Biotechnology
Research Areas
My laboratory will be focused on developing and applying enabling genomic technologies to decode the complex genetic networks underlying novel functions of biological systems and the molecular underpinnings of human diseases and aging. I have a multidisciplinary background across different biological and biomedical divisions with training and expertise in genome instability, genome engineering, cancer biology, synthetic biology, yeast genetics, plant biotechnology, biochemistry, systems biology and virology. The unique approach of my laboratory is to integrate genome engineering, biomedical science and synthetic biology through systematic dissection and modulation of cellular DNA repair and RNA regulation processes in diverse experimental models. The aims are to (i) develop new and advanced genome engineering technologies to facilitate biomedical research, (ii) better understand genome instability in human diseases for their prevention, early diagnostics and treatment, (iii) develop new precision medicine toolsets for cancer immunotherapy and gene therapy, and (iv) engineer new synthetic biology platforms for discovery and biosynthesis of therapeutic products.
Research directions:
1. Next-generation genome editing technology development and precision medicine application
2. Molecular mechanisms of genome instability in cancer, aging and diseases
3. Natural pharmaceutical product discovery and biosynthesis via synthetic biology platforms
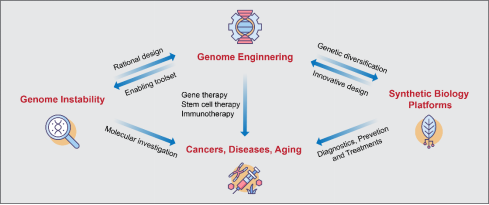
The interconnecting network of research areas in the Liang laboratory with an aim to promote basic biomedical science and translational clinical applications.
Highlights
Dr. Liang has hosted or participated in multiple research projects with about 1 million US dollars in accumulated funding in recent years. He published a number of high-profile research articles, in journals such as Nature Communications, Nucleic Acid Research, Cell Report, Plant Physiology as the first author or co-corresponding author; in Nature Biotechnology, ACS Central Science, Nature Communicates, PLOS Genetics, Journal of Biochemistry as co-authors. His primary contributions in science are as follows:
1. Unveiled new insights of plant DNA repair on the Agrobacterium transferred DNA (T-DNA) during plant genetic transformation.
2. Developed a new T-DNA vector system to facilitate construction of metabolic pathways and led to the first complete biosynthesis of potent anti-malarial drug artemisinin in a transgenic tobacco.
3. Developed a suite of new experimental methods to study NHEJ kinetics and fidelity, shedding light into the mechanism DNA double strand break (DSB) repair pathway choice and the mutagenic outcome.
4. Uncovered the link between RNA transcription and the Fanconi Anemia DNA interstrand crosslinking (ICL) repair protein FANCI-FANCD2 activation.
5. Improved greatly the editing efficiency and accuracy of the genome engineering technique‘eMAGE’and used it to study driver mutations of cancer relevant receptor proteins.
6. Developed a set of rapid and safe genomics tools to study SARS-CoV-2 pangenome mutations to facilitate pathogenesis study of COVID-19, high-throughput drug screening and renewal of vaccine to address emerging virus mutations.
Honors
• 2003 National First-Class Athlete of China (Chess)
• 2005 SJTU Excellent Academic Scholarship (A class – Top tier)
• 2007 SJTU Excellent League Cadres (Chairman of College Student Union and Founding Director of Student Science and Technology Innovation Center)
• 2008 Shanghai City College Fellowship
• 2008 Shanghai Excellent College Graduate
• 2015 Mary Sue and Kenneth Coleman Graduate Fellowship (UMich Presidential Award)
• 2015 Rackham Graduate Dissertation Fellowship
• 2016 Damon Runyon Postdoctoral Fellowship Award
• 2018 Yale Cancer Center Innovation Award
• 2020 Yale COVID-19 Pilot Award
Related News
1. UMich Student Spotlight (https://rackham.umich.edu/discover-rackham/student-spotlight-zhuobin-liang/)
2. Damon Runyon Cancer Research Foundation (https://www.damonrunyon.org/our-impact/current-projects/scientists/2351)
3. Awarded NSF-EDGE funding to develop cutting-edge genomics tools – as Co-PI in the Isaacs laboratory (https://nsfgov.home.blog/2019/09/17/the-cutting-edge-of-genomics/)
4. Yale COVID-19 Pilot Projects – as Co-PI in the Isaacs laboratory (https://covid.yale.edu/news-article/25722/)
Selected Publications
1. Liang Z.*, Metzner E. and Isaacs F.J.* (2020). “Advanced eMAGE for highly efficient combinatorial editing of a stable genome.” bioRxiv doi.org/10.1101/2020.08.30.256743 (Nat Commun, in revision) (IF=12.1)
2. Liang Z., Liang F., Teng Y., Chen X., Liu J., Longerich S., Rao T., Green A. M., Collins N. B., Xiong Y., Lan L., Sung P. and Kupfer G. M. (2019). “Binding of FANCI-FANCD2 complex to RNA and R-Loops stimulates robust FANCD2 monoubiquitination.” Cell Rep (IF=8.1)
3. Liang Z., Sunder S., Nallasivam S. and Wilson T. E. (2016). “Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining.” Nucleic Acids Res 44(6): 2769-2781. (IF=11.5)
4. Liang Z. and Tzfira T. (2013). “In vivo formation of double-stranded T-DNA molecules by T-strand priming.” Nat Commun 4: 2253. (IF=12.1)
5. Liang Z.*, Chiruvella K. K.*, and Wilson T. E. (2013). “Repair of double-strand breaks by end joining.” Cold Spring Harb Perspect Biol 5(5): a012757 – Chapter XVI of the Book: DNA Repair, Mutagenesis, and Other Responses to DNA Damage. (*co-first author)