Success is built on accumulation and fermentation of many failures
Zhou, Yaoqi
周耀旗
Institute of Systems and Physical Biology
ISPB Vice Director ,Senior Principal Investigator
zhouyq@szbl.ac.cn
Home page of research group:http://zhouyq-lab.szbl.ac.cn/
Timeline
-
2021~Present
Shenzhen Bay Laboratory, Shenzhen, China Senior Principal Investigator
-
2013~2021
Griffith University, QLD, Australia Professor
-
2006~2013
Indiana University Purdue University at Indianapolis, Indiana, USA Tenured Full Professor
-
2004~2006
State University of New York at Buffalo, USA Tenured Associate Professor
-
2000~2004
State University of New York at Buffalo, USA Assistant Professor
-
1995~2000
Harvard University, MA, USA Research Associate
-
1994~1995
North Carolina State University, NC, USA Postdoctoral Fellow
-
1994~1994
State University of New York at Stony Brook, NY, USA Postdoctoral Fellow
-
1990~1993
Applied P & Ch Laboratory, CA, USA Scientist and Director
Research Areas
Dr Zhou’s group focuses on the fundamental research in the relation between sequence, structure, and function of RNA and proteins as well as applied research in biomolecules. Our unique capability of combining structural bioinformatics and high-throughput technique with directed evolution biotechnology allows us to achieve the goals of deep mechanistic understanding of sequence, structure and function relation and improved application of biomolecules in precision medicine and detection of personalised biomarkers.
Highlights
Dr. Yaoqi Zhou obtained a bachelor's degree in chemical physics from the University of Science and Technology of China, a doctoral degree in computational statistical mechanics from the State University of New York at Stony Brook, and was a NIH postdoctoral fellow in computational biology at Harvard University. He was an assistant professor at the State University of New York at Buffalo in 2000, promoted to a tenured associate professor in 2004, a professor in the Schools of Informatics and Medicine of Indiana University in 2006. He became a professor and principal research leader at the Institute of Glycomics, Griffith University, Australia in 2013. Currently, he is a senior principal investigator at the Institute for System and Physical Biology, Shenzhen Bay Laboratory, Shenzhen, China. His extensive and in-depth scientific training enables him to lead doctoral and postdoctoral teams during his work in the United States, Australia, and China, and to collaborate with multidisciplinary researchers in bioinformatics and its applications in molecular biology. His unique and original contributions include the concept that the native state of proteins is a surface molten solid, development of an all-atom potential function DFIRE based on statistical mechanics, and the template-based protein tertiary structure prediction method SPARKS. He was the first to apply deep learning methods to predict the structure of protein backbones (SPIDER/SPOT-1D) and RNA secondary structure (SPOT-RNA). He designed new peptide drugs that cells will not be able to develop resistance by disrupting the structure of a target protein, and inferred the structure of RNA through high-throughput sequencing. His group has repeatedly ranked among the best in international protein-structure prediction and function prediction competitions (first place in the blind prediction of protein structures in the CASP6-2004 server group, first place in CAGI-2013 blind prediction of cell proliferation rate, first place in CAID-2019 blind prediction of protein intrinsic disorder). Dr Zhou has received multiple funding from the NIH of the United States, the NHMRC and ARC of Australia, and the key research and development projects of Guangdong Province, and etc. He has published more than 200 papers in Genome Biology, Ann. Review Biophysics, PNAS, Nucleic Acids Research, Cell Metabolism, Nature Communications, Nature Machine Intelligence and other international well-established journals. According to Google Scholar, his work has a total of more than 13,000 citations with an H-index of 61 and more than 1,300 citations in 2020.
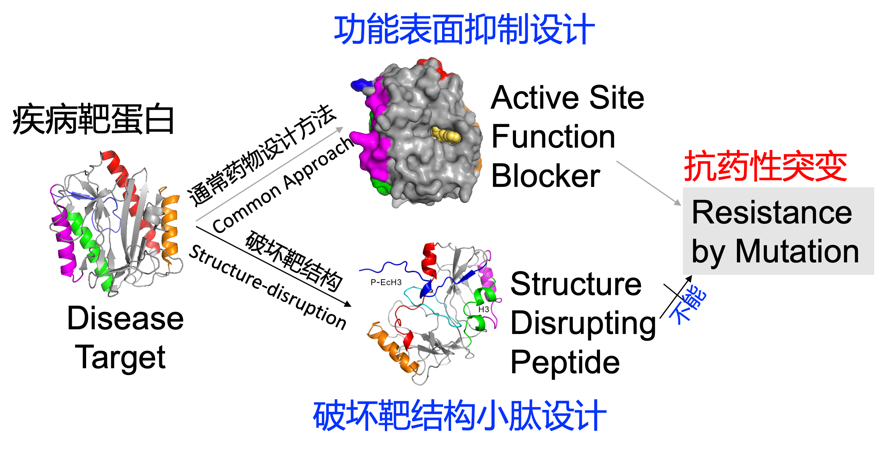
1. Most drugs were designed to block the function of a target protein by sticking onto its functional surface. Such a design often leads to drug resistance by spontaneous random mutation of the target. Dr. Zhou has designed peptides to disrupt the structure of a target protein so that the bacterial or cancer cells will not be able to develop resistance by mutations (Zhan and Zhou et al. FASEB J. , 33: 2095–2104 (2019);News release, http://www.china.org.cn/world/Off_the_Wire/2018-09/28/content_64238761.htm).

2. The first end-to-end prediction of RNA secondary structure significantly improved the accuracy of prediction (Nature Communications 10, 5407 (2019)) News (https://www.brisbanetimes.com.au/national/queensland/researchers-use-ai-deep-learning-to-predict-human-rna-structures-20191127-p53er1.html)
Honors
• 1985, Sino-American Chemistry Graduate Program (CGP)
• 1989, The Lee Meyer Outstanding Graduate Student Award, Chemistry Department, State University of New York at Stony Brook, USA.
• 1995-1996, Postdoctoral fellowship award, NSF Program in Mathematics and Molecular Biology
• 1996-1998,NIH Postdoctoral fellowship award
• 2006, Henry A. Panasci Jr. Technology Entrepreneurship Competition 2006 CTO, BioHammer
• 2012, Excellence in Research Award, School of Informatics, Indiana University School of Informatics, Indiana, USA.
• 2018, Excellence in Research Leadership Award, Institute for Glycomics,Griffith University, QLD, Australia
Selected Publications
1. J. Singh, J. Hanson, K. Paliwal, and Y. Zhou, RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning, Nature Communications 10, 5407 (2019).
2. Z. Zhang, P. Xiong, T. Zhang, J. Wang, J. Zhan, and Y. Zhou, Accurate inference of the full base-pairing structure of RNA by deep mutational scanning and covariation-induced deviation of activity, Nucleic Acids Research, 48:1451-1465 (2020).
3. J. Zhan, H. Jia, E. A. Semchenko, Y. Bian, A. M. Zhou, Z. Li, Y. Yang, J. Wang, S. Sarkar, M. Totsika, H. Blanchard, F. E.-C. Jen, Q. Ye, T. Haselhorst, M. P. Jennings, K. L. Seib, and Y. Zhou, Self-derived structure-disrupting peptides targeting methionine aminopeptidase in pathogenic bacteria; a new strategy to generate antimicrobial peptides, FASEB J. , 33: 2095–2104 (2019).
4. S. Xu, J. Zhan, B. Man, S. Jiang, W. Yue, S. Gao, C. Guo, H. Liu, Z. Li, J. Wang, and Y. Zhou, Real-time reliable determination of binding kinetics of DNA hybridization using a multi-channel graphene biosensor, Nature Communications 8, 14902 (2017).
5. Z. Li, Y. Yang, J. Zhan, L. Dai and Y. Zhou, Energy Functions in De Novo Protein Design: Current Challenges and Future Prospects, Ann. Rev. Biophysics 42, 315-335 (2013).