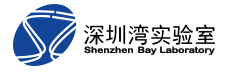Chance favors the prepared mind!
Peng, Qin
彭琴
Institute of Systems and Physical Biology
Junior Principal Investigator
pengqin@szbl.ac.cn
Home page of research group:http://pengqin.szbl.ac.cn/
Timeline
-
2020 - Present
Shenzhen Bay Laboratory Junior Principal Investigator
-
2017 - 2020
UC San Diego Assistant Project Scientist
-
2015 - 2017
UC San Diego Postdoctoral Researcher
-
2010 - 2015
Chongqing University / University of Illinois, Urbana-Champaign & University of California, San Diego PhD/Joint PhD
-
2006 - 2010
Chongqing University Bachelor
Research Areas
The main goal of Dr. Peng’s research is to integrate different cutting-edge technologies, including Fluorescence Resonance Energy Transfer (FRET), molecular engineering, live cell imaging, chromatin labeling and biomechanics, to develop epigenetic FRET biosensors in order to address fundamental biological/physiological questions, e.g. How do mechanical cues regulate chromatin and epigenetics not only in global but also at specific genomic loci. Her group will focus on the following projects:
1. Nuclear mechanics regulated epigenetics;
2. Development of 4D imaging platform of epigenetics;
3. Regulation of nuclear mechanics in chromatin/epigenetics in transdifferentiation.
Highlights
Histone modifications change DNA accessibility to either activate or suppress genes, resulting in different diseases, e.g. cancer. It is hence important to have mechanistic understanding of epigenetic regulations. The main goal of my research is to integrate different cutting-edge technologies, including Fluorescence Resonance Energy Transfer (FRET), molecular engineering, live cell imaging, chromatin labeling and biomechanics, to develop epigenetic FRET biosensors in order to address fundamental biological/physiological questions, e.g. epigenetic regulation and mechanotransduction, not only in global but also at specific genomic loci. I mainly studied on real-time coordination among different histone modifications in regulating cellular functions - for example, the histones H3K9me3 and H3S10ph. Visualizing this coordination can allow us to identify key epigenetic markers in governing chromatin structure and duration of the cell cycle. These FRET biosensors can hence be used to screen specific drugs to treat cell cycle-associated diseases. Additionally, I conducted a high-throughput screening method by using a yeast display system to screen and identify monobodies (single-domain synthetic binding proteins). These monobodies recognize target molecules, allowing the assembly of FRET biosensor at cell surface to monitor extracellular membrane enzymatic activities. They can also be applied to target histone modifications with high specificity and affinity for FRET biosensor engineering to monitor various histone modifications in the future. Last but not the least, I developed a new system for genomic loci labeling and epigenetic manipulation in single live cells based on phase separation, which will lead to our in-depth understanding of the role of epigenetic modifications in genomic regulation in response to physical and chemical environmental cues.
I have published 23 papers including PNAS, CELL CHEM, SCI ADV, CURR OPIN SOLID ST M, J MOL BIOL, and so on. I have invited to present my work on BMES, 4DN, ICBME conferences and shared my biosensors with many other laboratories all over the world, e.g. Harvard medical school, Cornell University, UCLA, UCI, etc. Based on my work, I became the main contributor and project leader of four major NIH grants that we have received in the previous lab (R33CA204704A (Multiplex FRET Imaging of Kinase-Epigenome Interregulations in Live Cancer Cells), R01 GM125379-01 (Locus-specific Imaging of Dynamic Histone Methylations during Reprogramming), 5R01HL121365-06 (Role of Spatiotemporal Epigenetic Dynamics in Regulating Endothelial Gene Expressions under Flows), 1R01GM126016-01 (Remote-Control Mechano-Genetics and Epigenetics for Live Cell Manipulation)). I have also served as a reviewer to various journals as well as providing reviewing feedback to grant applications to the National Science Foundation (NSF) and National Institute of Health (NIH).
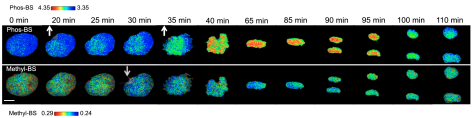
Fig 1. Dual-FRET Imaging Simultaneously Visualizes H3K9me3 and H3S10ph
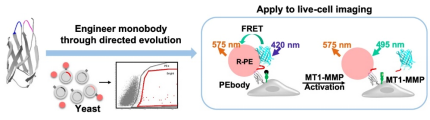
Fig 2. The universal platform for different kinds of epigenetic FRET biosensor generation and optimization
Honors
• 2016 UCSD Postdoc Travel Award
• 2012-2014 China Scholarship Council (CSC) Joint Ph.D. Program
• 2010 Excellent Graduation Project of Chongqing University
• 2009 Huang Qianheng Foundation Scholarship
• 2008 Excellent Student of Chongqing University
Related News
https://www.szbl.ac.cn/infomation/topic/interview/977.html
Selected Publications
[1] Peng Q. *, †, Huang Z. *, Sun K., Liu Y., Yoon C., Harrison R., Schmitt D., Zhu L., Wu Y., Tasan I., Zhao H., Zhang J., Zhong S., Wang Y†. Engineering Inducible Biomolecular Assemblies for Genome Imaging and Manipulation in Living Cells. Nat Commun, 2022, 13(1): 7933. DOI: 10.1038/s41467-022-35504-x. († Corresponding author, Featured Article)
[2] Harrison R., Weng K., Wang Y.†, Peng Q.†. Phase Separation and Histone Epigenetics in Genome Regulation. Curr Opin Solid St M. 2021, 25(1):100892. doi: 10.1016/j.cossms.2020.100892. († Corresponding author)
[3] Huang Z, Ouyang M., Lu S., Wang Y†, Peng Q.†. Optogenetic Control for Investigating Subcellular Localization of Fyn Kinase Activity in Single Live Cells. J Mol Biol. 2020, pii: S0022-2836(20)30236-9. DOI: 10.1016/j.jmb.2020.03.015. († Corresponding author)
[4] Peng Q., Lu S., Shi Y., Pan Y., Limsakul P., Chernov AV., Qiu J., Chai X., Shi Y., Wang P., Ji Y., Li YJ., Strongin AY., Verkhusha VV., Izpisua Belmonte JC., Ren B., Wang Y., Chien S. †, Wang Y†. Coordinated Histone Modifications and Chromatin Reorganization in a Single Cell Revealed by FRET Biosensors. Proc Natl Acad Sci U S A. 2018, 115(50): E11681-E11690. doi: 10.1073/pnas.
[5] Limsakul P. *, Peng Q. *, Wu Y., Allen ME., Liang J., Remacle AG., Lopez T., Ge X., Kay BK., Zhao H., Strongin AY., Yang XL., Lu S., Wang Y †. Directed Evolution to Engineer Monobody for FRET Biosensor Assembly at Live-Cell Surface. Cell Chem Biol. 2018, 25(4): 370-379.e4. doi: 10.1016/j.chembiol.2018.01.002. * Equal contribution.