Yin, Yandong
尹延东
Institute of Chemical Biology
Junior Principal Investigator
yinyd@szbl.ac.cn
Timeline
-
2021 - Present
Shenzhen Bay Laboratory Junior Principal Investigator
-
2018 - 2021
New York University School of Medicine Research Scientist
-
2014 - 2018
New York University School of Medicine Postdoc Fellow
-
2013 - 2014
Peking University Research Assistant
-
2008 - 2013
Peking University PhD
-
2004 - 2008
Peking University BS
Research Areas
The Yandong Group is focusing on innovating biophysical technologies in Super-Resolution Imaging, Image Deep Mining, and live-cell Single-Molecule Tracking. The major interest is to understand the biological physics at single-molecule level by integrating the state-of-the-art techniques in optical imaging, data mining, and chemical biology.
Highlights
I. High-Content Super-Resolution Image Analysis
The multi-color Super-Resolution Microscopy enabled visualization of molecular complex at a scale of 5-15 nm, it is however yet widely applied due to the challenges in deriving quantitative information from the complex image data. Dr. Yandong Yin developed new Stochastic Co-localization modeling and Triple-Correlation (TC) based image mining method that provides higher order statistical information on the spatial organization of biomolecular complexes. In addition, Dr. Yandong Yin also developed fast computational method addressing the expensive TC analysis, enabling high-throughput high-content super-resolution image analysis (Nat. Commun., 2019).
Utilizing these quantitative microscopy methods, Dr. Yandong Yin investigated 1) the molecular mechanism of ATR modulating FANCI’s activity in responding to different level of DNA replication stress (Mol. Cell, 2015), and 2) quantified the spatial organization of individual replisome at single-fork resolution, revealing the physical link between the ATR-mediated surveillance of individual replication fork and stress response (Mol. Cell, 2021).
II. Diffusion-Decelerated Fluorescence Correlation Spectroscopy
Single-molecule techniques could be ideal in probing subtle conformational dynamics, but existing single-molecule techniques had yet cover full temporal window for kinetic studies. Dr. Yandong Yin developed the diffusion-decelerated FCS (ddFCS), enabling a much wider time window for measurements of the dynamic of subtle conformational changes at single-molecule level. As such, Dr. Yandong Yin mapped the panorama of the folding kinetics of the DNA hairpin structure (Chem. Commun., 2012), and probed, for the first time, the dynamics of a single mismatched base spontaneously flipping out of its duplex structure (PNAS, 2014).
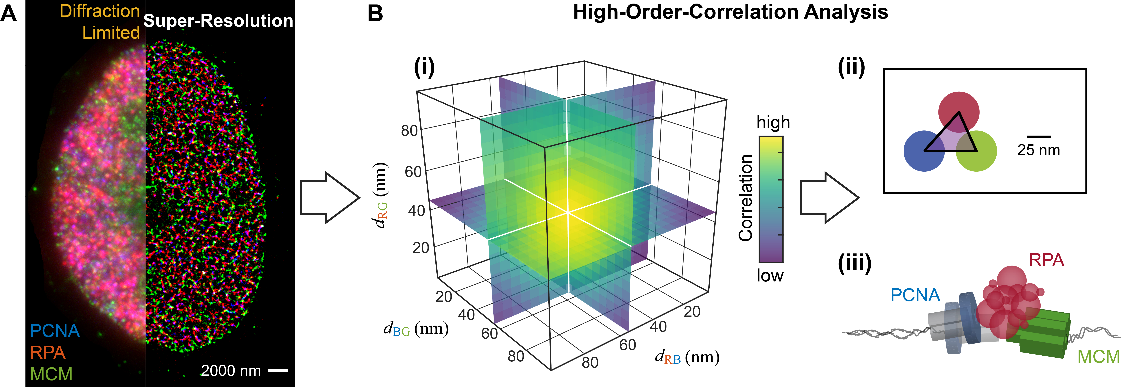
Figure 1. High-Content Super-Resolution Image Mining
(A) Multi-Color Super-Resolution imaging provides nanoscale spatial information. (B) The Triple-Correlation Function (TCF) quantifies the spatial organization of the molecular complex: (i) The TCF plots the relative probability density as a function of the pairwise distances of the three examined species; (ii) The most probable configuration obtained from the TCF profile; (iii) Quantification of the spatial organization of the molecular complex.
Honors
• 2013 Outstanding Doctoral Dissertation of Peking University, Peking University
• 2012 National Scholarship for Outstanding Graduate Student of China
• 2012 Celanese Scholarship, Peking University
Publication
1. Y Yin#, WTC Lee, D Gupta, …, E Rothenberg#, A basal-level activity of ATR links replication fork surveillance and stress response, Mol. Cell 2021, 81, 4343-4257 (#corresponding author).
2. WTC Lee, Y Yin, M Morton, …, E Rothenberg, Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling, Nat. Commun. 2021, 12, 1-14.
3. Y Yin#, WTC Lee, E Rothenberg#, Ultrafast data mining of molecular assemblies in multiplexed high-density super-resolution images, Nat. Commun. 2019, 10, 119 (#corresponding author).
4. YH Chen*, MJK Jones*, Y Yin*, SB Crist, …, TT Huang, ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress, Mol. Cell 2015, 58, 323-338 (*co-first author).
5. Y Yin*, L Yang*, G Zheng, C Gu, C Yi, C He, Y Gao, XS Zhao, Dynamics of spontaneous flipping of a mismatched base in DNA duplex, Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 8043-8048 (*co-first author)