Research teams at the Institute of Molecular Physiology at Shenzhen Bay Laboratory and the Genome Institute of Singapore (GIS) have collaborated together to develop xPore, a software that extracts RNA modifications (an additional layer of information above the genetic molecule RNA) from transcriptomics data. Their research was published in Nature Biotechnology on July 19, 2021.
"Imagine typing the same word twice in one line. While both words can have the same spelling, differences in how the words are formatted (e.g. underlined or in italics) can change how we emphasis each word. Likewise, chemical modifications to 1 RNA may change how it functions compared to an unmodified RNA with the same underlying RNA sequence.” said Dr. Sho Goh, Assistant Professor from Shenzhen Bay Laboratory who co-led the study. “These RNA modifications are widespread, but because they do not change the letters of the RNA, they are very difficult to identify," said Dr Jonathan Göke, Group Leader of Laboratory of Computational Transcriptomics at GIS who also co-led the study. More than 100 RNA modifications are known to play different roles in cells. Some of these RNA modifications are associated with disease risk, while others are used in mRNA vaccines. In the past, identifying RNA modifications required labour- and time-intensive bench-experiment assays that only very few laboratories can perform.
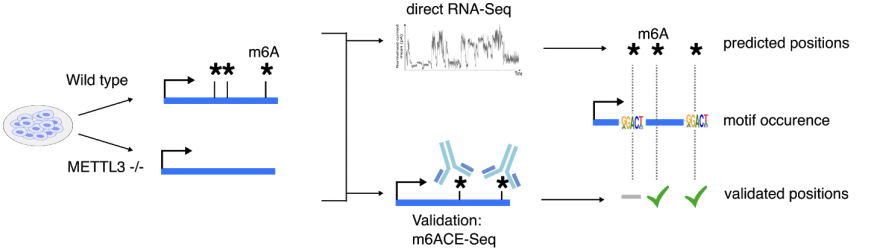
Development of xPore algorithm for predicting m6A positions from direct RNA-seq data.
To overcome these limitations, the team utilised direct Nanopore RNA sequencing, a new transcriptomics technology that sequences a raw RNA molecule with its modifications retained. To extract the hidden layer of RNA modifications, they developed xPore, a machine learning-based method that precisely detects differences in RNA modifications. A property employed in the method is the consistent data of the unmodified sites, and the existence of modifications disrupts this consistency.
"Similar problems occur in other data-rich areas such as finance or speech recognition. Here, we adopted an existing statistical model that is used frequently in data science, so that it can precisely identify these modified sites," explained Dr Ploy Pratanwanich, previous Postdoctoral Fellow at GIS and the first author of the study. In their study, the authors demonstrated that xPore is highly accurate and is able to overcome many of the previous limitations to studying RNA modifications.
Dr. Goh added, “The ability to map new RNA modifications is vital for determining their functions. Since xPore does not require specific reagents that specialise in identifying only a single RNA modification type, it can potentially detect other RNA modifications beyond m6A. Therefore, xPore’s flexibility can expedite our efforts to discover novel RNA modification functions.”
吴炜祥课题组与合作者发表研究:Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore
深圳湾实验室分子生理学研究所吴炜祥课题组和新加坡基因组研究所(GIS)的研究团队合作开发了xPore软件,它可以从基因组数据中筛选出含有RNA修饰(RNA上的附加信息层)的相关信息。该研究于2021年7月19日在著名期刊《自然生物技术》(Nature Biotechnology)上发表。
吴炜祥博士举例说,“对于在同一文本中的两个完全一样的单词,我们可以通过对其设置不同的格式(如下划线或斜体)来体现我们对每个单词不同的强调方式。同样地,在RNA序列相同的前提下,与未经修饰的RNA相比,经过化学修饰的RNA可能会改变其功能”。GIS计算转录组学实验室组长Dr Jonathan Göke指出,“RNA修饰普遍存在,但其并不改变原有基因序列,因此难以被识别。”目前已知有100多种RNA修饰在细胞中发挥着不同的作用。其中一部分与疾病风险有关,另一部分则被用于mRNA疫苗上。但是识别RNA修饰需要耗费实验室大量的人力及时间成本,因此只有少数实验室能开展这项工作。
研究团队采用了一种新的转录组学技术以解决目前的难题,即纳米孔直接RNA测序,它可以在保留RNA修饰的基础上对其进行测序。为了提取RNA修饰的隐藏层,研究团队开发了一种基于机器学习的方法--xPore,它可以精确检测出RNA修饰的差异。该方法的主要依据是未修饰位点的一致性数据,只要存在RNA修饰该数据的一致性就会被打破。
“类似的问题也出现在其他数据丰富的领域,如金融或语音识别。本研究中,我们采用了数据科学中现有的常用统计模型,以便精确地识别这些被修饰过的位点”,GIS的前博后研究员Ploy Pratanwanich(该文章第一作者)解释说。其研究证明xPore是高度精确的,并且能够克服以往研究RNA修饰时存在的许多限制。
吴炜祥博士补充道,“绘制RNA修饰图的能力对确定它们的功能至关重要。由于xPore不需要专门鉴定单个RNA修饰类型的特定试剂,所以它有可能检测到除m6A以外的其他RNA修饰。因此,xPore的灵活性可以推动功能性RNA修饰领域的发展” 。

特聘研究员
The Goh lab studies how RNA modifications (epitranscriptome) regulate gene expression and cellular processes by developing new technologies to sequence RNA modifications and functionally characterizing novel epitranscriptomic factors that interact with and metabolize RNA modifications. Defects in epitranscriptomic regulation cause a variety of diseases and determining the mechanistic basis that underlie these defects will be crucial for developing effective epitranscriptomic-based therapeutic strategies.
In recent years, Sho has focused on the study of RNA modifications, also known as Epitranscriptomics. One of his key contributions to this field is the development of high-resolution epitranscriptome sequencing technologies. These technological advancements helped facilitate the discovery of novel epitranscriptomic factors that generate RNA modifications, which revealed new mechanisms by which modified RNA is regulated. Sho has also challenged popularly held notions of epitranscriptomic pathways by using precise sequencing methods to accurately identify the metabolic targets of previously known epitranscriptomic factors and thus, redefined their functional roles. As a member of the RNA society since 2020, Sho's work has been published in prestigious journals (including Nature Biotechnology, Nature Communications, Nucleic Acids Research, Genes & Development, PLoS Computational Biology and Cell ) and resulted in a patent and multiple research grants.
论文标题:
Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore
论文全文:
https://www.nature.com/articles/s41587-021-00949-w
文章来源 | 吴炜祥课题组
编辑 | 鲍 啦