
电话:+86-755-86967710
邮箱:webmaster@szbl.ac.cn

系统与物理生物学研究所
artem@szbl.ac.cn
物理生物学
Junior Principal Investigator
Senior postdoctoral research fellow
Postdoctoral research fellow
Postdoctoral research fellow
Undergraduatestudy, Graduate study, Ph.D.
1. Study of molecular mechanisms responsible for mechanotransduction in living cells.
2. Understanding the role of mechanical and topological constraints in regulation of DNA-protein interactions and chromatin organization in living cells.
3. Exploration of molecular mechanisms underlying mechanochemical coupling in cytoskeletal motor proteins, and their role in regulation of intracellular transportation.
4. Development and application of single-molecule force-spectroscopy methods for investigation of the above questions.
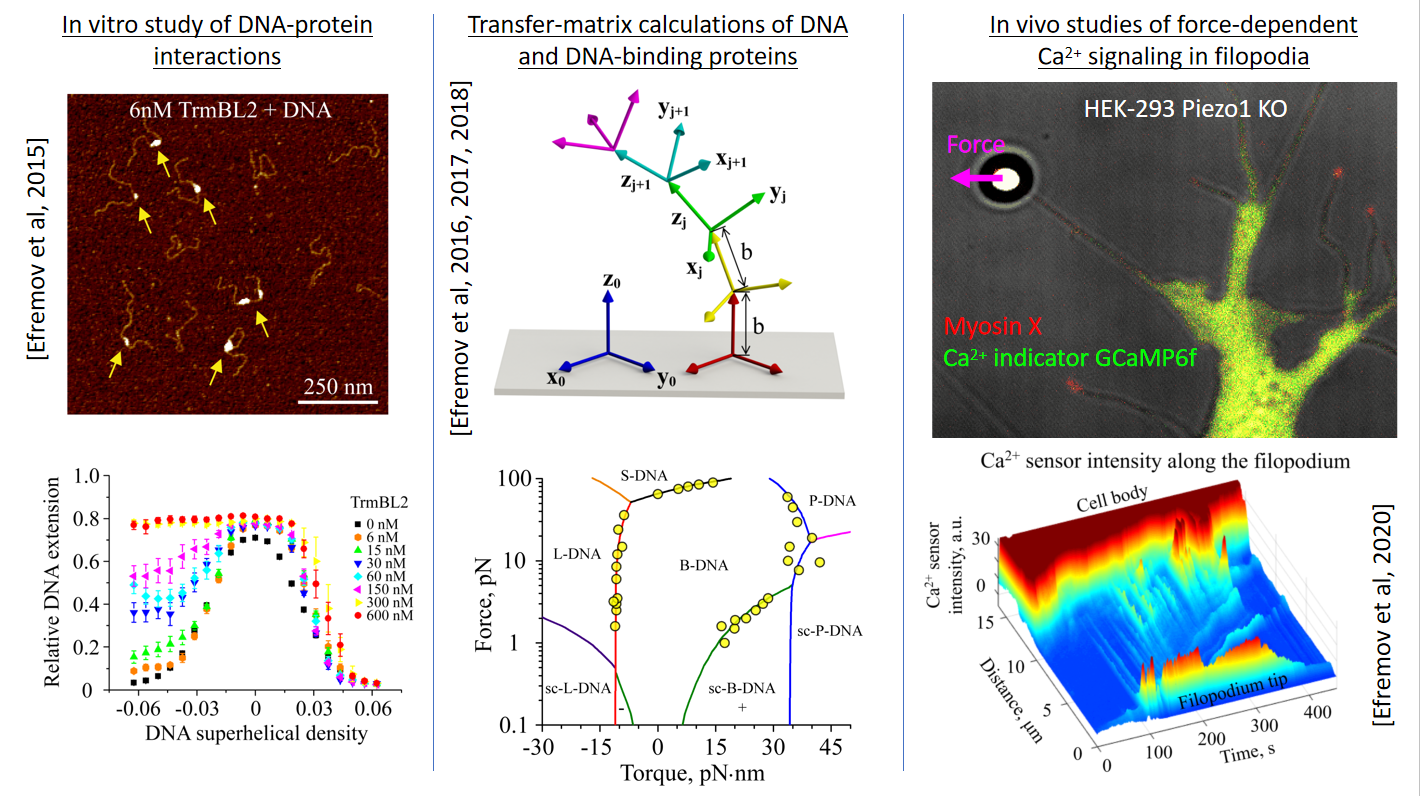
During past several years Artem has been applying various experimental and theoretical biophysical methods, such as optical and magnetic tweezers, confocal and atomic force microscopy as well as elements of chemical kinetics and quantum physics theories, to explore different biological systems at molecular level ranging from kinesin and myosin motor proteins to DNA-architectural complexes. These researches done in collaboration with world leading biophysical groups led to several interesting discoveries and practical implications such as construction of light-driven nano-motors and development of experimental assays, which can be used for investigation of human-related neural and parasitic diseases at single-molecule level. Artem’s work has been published in many high impact journals including Cell, PNAS, Nature Communications and Nucleic Acids Research. He has been a member of American Biophysical Society and Australian Society for Biophysics since 2015 and 2017, respectively.
1.Efremov A K, Yao M, Sun Y, et al. Application of piconewton forces to individual filopodia reveals mechanosensory role of L-type Ca2+ channels[J].Biomaterials, 2022, 284: 121477.
2. Alieva N O,Efremov A K, Hu S, et al. Myosin IIA and formin dependent mechanosensitivity of filopodia adhesion[J].Nature communications, 2019, 10(1): 1-14.
3.Efremov A K, Yan J. Transfer-matrix calculations of the effects of tension and torque constraints on DNA–protein interactions[J].Nucleic acids research, 2018, 46(13): 6504-6527.
4.Efremov A K, Radhakrishnan A, Tsao D S, et al. Delineating cooperative responses of processive motors in living cells[J].Proceedings of the National Academy of Sciences,2014, 111(3): E334-E343.
5. Xu X,Efremov A K, Li A, et al. Probing the cytoadherence of malaria infected red blood cells under flow[J].PloS one, 2013, 8(5): e64763.
6.Efremov A K, Grishchuk E L, McIntosh J R, et al. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions[J].Proceedings of the National Academy of Sciences, 2007, 104(48): 19017-19022.
7. Grishchuk E L,Efremov A K, Volkov V A, et al. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion[J].Proceedings of the National Academy of Sciences, 2008, 105(40): 15423-15428.
8.Efremov A K, Qu Y, Maruyama H, et al. Transcriptional repressor TrmBL2 from Thermococcus kodakarensis forms filamentous nucleoprotein structures and competes with histones for DNA binding in a salt-and DNA supercoiling-dependent manner[J].Journal of Biological Chemistry, 2015, 290(25): 15770-15784.
9. Lu H,Efremov A K, Bookwalter C S, et al. Collective dynamics of elastically coupled myosin V motors[J].Journal of Biological Chemistry, 2012, 287(33): 27753-27761.
10.Efremov A K, Winardhi R S, Yan J. Transfer-matrix calculations of DNA polymer micromechanics under tension and torque constraints[J].Physical Review E,2016, 94(3): 032404.
Personal Websites
1) Google Scholar:https://scholar.google.com/citations?user=DHS_uKEAAAAJ&hl=en
2) Researchgate:https://www.researchgate.net/profile/Artem_Efremov
联系方式
地址:深圳市光明区高科创新中心
电话:+86-755-86967710
邮箱:webmaster@szbl.ac.cn
邮编:518132
Copyright © 2025 深圳湾实验室 版权所有www.szbl.ac.cn All Rights Reserved.粤ICP备19032884号 粤公网安备44031102000926号
粤公网安备44031102000926号


