
电话:+86-755-86967710
邮箱:webmaster@szbl.ac.cn
特聘研究员
研究助理教授
博士后
博士
课题组实验和计算相结合,科研与临床相结合,注重转化,研究方向包括但不限于:
1. 癌症早期筛查、诊断(Cancer Liquid Biopsy)
2. 肿瘤转移的分子机理与抑制方法(Cancer metastasis)
3. 长链非编码RNA功能(Long noncoding RNA)
4. 多组学技术、生物信息学(Multi-omics/Bioinformatics)
5. 人工智能技术应用(Artificial Intelligence)
更多信息请访问https://hellosunking.github.io/,课题组招聘助研/博士后/联培博士研究生/科研助理等岗位,岗位投递请发送简历至sunkun@szbl.ac.cn或添加课题组微信(Sunkunlab)进行联系。
孙坤博士长期从事生物信息学和基于外周血游离DNA进行癌症诊断的研究,发表SCI文章50余篇,其中(共同)第一作者或通讯作者30余篇(包括Cancer Discovery、Genome Research、Nature Communications、PNAS、EMBO Journal等),总被引用5100余次。孙坤博士主持多项国家级科研项目,包括国家级青年人才项目、国家重点研发计划(首席科学家)等。孙坤博士的工作注重原创性和转化价值,申请了15+项国际、国内专利,部分专利均已成功转化落地。
孙坤课题组计算与实验并重,自成立以来以深圳湾实验室为第一或通讯作者身份已经发表了10余篇SCI论文,包括Nature Communications2023、Cell Reports Methods2024a, 2024b、Briefings in Bioinformatics2021、Patterns2020、Bioinformatics2020等。
详细介绍请查看:https://hellosunking.github.io。
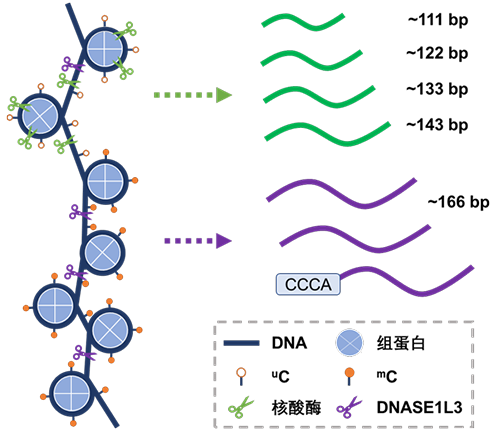
揭示游离DNA片段化模式的分子机理以及应用
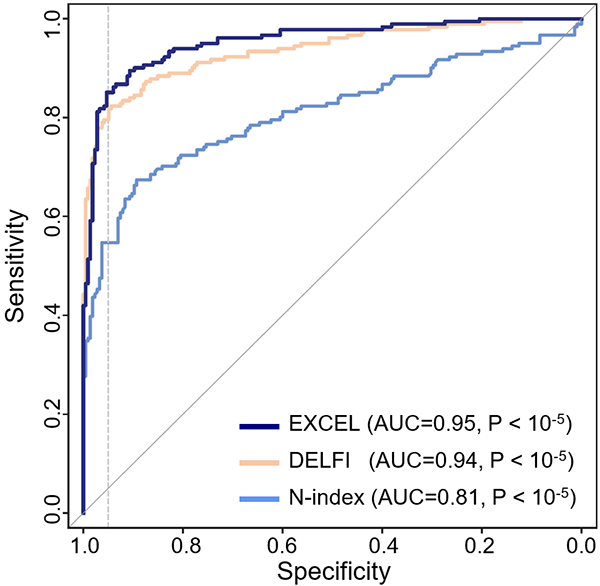
完全自主知识产权的高准确率、高敏感度癌症诊断大模型
1. An Y, Zhao X, Zhang Z, Xia Z, Yang M, Ma L, Zhao Y, Xu G, Du S, Wu X, Zhang S, Hong X, Jin X*,Sun K*. DNA methylation analysis explores the molecular basis of plasma cell-free DNA fragmentation.Nat Commun2023 Jan 18; 14(1):287.
2. Jiang P#,Sun K#, Peng W#, Cheng SH, Ni M, Yeung PC, Heung MMS, Xie T, Shang H, Zhou Z, Chan RWY, Wong J, Wong VWS, Poon LC, Leung TY, Lam WKJ, Chan JYK, Chan HLY, Chan KCA, Chiu RWK, Lo YMD. Plasma DNA end motif profiling as a fragmentomic marker in cancer, pregnancy and transplantation.Cancer Discov2020 May; 10(5):664-673.
3.Sun K*, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, Ng SSM, Ma BBY, Leung TY, Chan SL, Mok TSK, Lai PBS, Chan HLY, Sun H, Chan KCA, Chiu RWK, Lo YMD*. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin.Genome Res2019 Mar; 29(3):418-427.
4.Sun K, Jiang P, Wong AIC, Cheng YKY, Cheng SH, Zhang H, Chan KCA, Leung TY, Chiu RWK, Lo YMD. Size-tagged preferred ends in maternal plasma DNA shed light on production mechanism and show utility in noninvasive prenatal testing.Proc Natl Acad Sci U S A2018 May 29; 115(22):E5106-E5114.
5. Jiang P#,Sun K#, Tong YK, Cheng SH, Cheng THT, Heung MMS, Wong J, Wong VWS, Chan HLY, Chan KCA, Lo YMD, Chiu RWK. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma.Proc Natl Acad Sci U S A2018 Nov 13; 115(46):E10925-E10933.
6. Chan KCA#, Jiang P#,Sun K#, Cheng YKY, Tong YK, Cheng SH, Wong AIC, Hudecova I, Leung TY, Chiu RWK, Lo YMD. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance and preferred DNA ends.Proc Natl Acad Sci U S A2016 Dec 13; 113(50):E8159-8168.
7.Sun K#, Jiang P#, Chan KCA#, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, Chan RW, Tong YK, Ng SS, Mong RSM, Hui DS, Leung TN, Leung TY, Lai PBS, Chiu RWK, Lo YMD. Plasma DNA tissue mapping by genomewide methylation sequencing for noninvasive prenatal, cancer and transplantation assessments.Proc Natl Acad Sci U S A2015 Oct 6; 112(40):E5503-12.
8.Sun K*, Wang H, Sun H*. NAMS webserver: coding potential assessment and functional annotation of plant transcripts.Brief Bioinform2021 May 20; 22(3):bbaa200.
9.Sun K*, Li L, Ma L, Zhao Y, Deng L, Wang H, Sun H. Msuite: a high-performance and versatile DNA methylation data-analysis toolkit.Patterns (N Y)2020 Oct 15; 1(8):100127.
10. Zhou L#,Sun K#, Zhao Y#, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, Zhu X, Wang L, Tang LY, Esteban MA, Wang R, Jauch R, Sun H, Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1.Nat Commun2015 Dec 11; 6:10026.
11. Lu L#,Sun K#, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis.EMBO J2013 Oct 2; 32(19):2575-88.
12. Chan KCA#, Jiang P#,Sun K#, Cheng YKY, Tong YK, Cheng SH, Wong AIC, Hudecova I, Leung TY, Chiu RWK, Lo YMD. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance and preferred DNA ends.Proc Natl Acad Sci U S A2016 Dec 13; 113(50):E8159-8168.
13.Sun K#, Jiang P#, Chan KCA#, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, Chan RW, Tong YK, Ng SS, Mong RSM, Hui DS, Leung TN, Leung TY, Lai PBS, Chiu RWK, Lo YMD. Plasma DNA tissue mapping by genomewide methylation sequencing for noninvasive prenatal, cancer and transplantation assessments.Proc Natl Acad Sci U S A2015 Oct 6; 112(40):E5503-12.
14. Zhou L#,Sun K#, Zhao Y#, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, Zhu X, Wang L, Tang LY, Esteban MA, Wang R, Jauch R, Sun H, Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1.Nat Commun2015 Dec 11; 6:10026.
15. Lu L#,Sun K#, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis.EMBO J2013 Oct 2; 32(19):2575-88.
联系方式
地址:深圳市光明区光侨路高科创新中心
电话:+86-755-86967710
邮箱:webmaster@szbl.ac.cn
邮编:518132
Copyright © 2024 深圳湾实验室 版权所有www.szbl.ac.cn All Rights Reserved.粤ICP备19032884号 粤公网安备44031102000926号
粤公网安备44031102000926号



